Biotek Gen5
Getting started with the Gen5 connector for the Agilent Biotek Gen5 software
This jupyter notebook introduces how the Gen5 connector is used with the UniteLabs SDK. The Gen5 connector is provided by UniteLabs. It is based on the UniteLabs CDK and is directly connected to the UniteLabs platform. The SiLA 2 interface can be used as is and implements a range of high-level commands. The UniteLabs SDK (extension) contains a range of further classes and methods related to this connector specifically. The Agilent Biotek Gen5 software connects to a wide range of plate readers from Agilent Biotek:
- ELx800
- ELx808
- Synergy HT
- FLx800
- Powerwave
- Clarity
- Synergy2
- PowerWaveXS 2
- Synergy Mx
- Epoch
- Synergy H4
- Synergy H1
- Eon
- Synergy Neo
- Cytation3
- Synergy HTX
- Cytation5
- Epoch 2
- Synergy Neo2
- Lionheart FX
- 800TS
- Cytation1
- Synergy LX
- Lionheart LX
- Cytation7
- LogPhase 600
- Cytation C10
The connector was developed and tested with Gen5 software version 3.14.
import asyncio
import json
from datetime import datetime, timezone
import matplotlib.pyplot as plt
import numpy as np
import csv
from IPython.display import clear_output
import nest_asyncio
nest_asyncio.apply()
import unitelabs
from unitelabs.sdk import Client
from unitelabs.sdk.core import ExecutionError
List all connected devices
Requests all connected connectors from the UniteLabs API
client = Client()
await client.list_services()
[
...,
Gen5Software(client=<unitelabs.sdk.client.client.Client object at 0x7f5ed9101a50>, id='<uuid>', name='Gen5 Software'),
...
]
Instantiate a Gen5 object
# Current option: get service by name
gen5 = await client.get_service_by_name('Gen5 Software')
# To make sure the command set is still the same, check the server version
await gen5.sila_service.get_server_version()
'1.0.1'
Explore the Gen5 modules
The Gen5 connector has 5 modules for different functionalities:
- sila_service: a standard suit of getters to retrieve information concerning the SiLA server
- gen5_service: all the low-level calls needed to process a plate using a previously established protocol
- door_controler: calls to get information about the carrier state and open and close the carrier
- temperature_controller: calls to manage the incubation in readers supporting temperature control.
- protocol_controller: currently supports creating a protocol with absorbance and/or shaker steps.
gen5.modules
{'sila_service': SiLAService(client=<unitelabs.sdk.client.client.Client object at 0x7f5ed9101a50>, id='9ee85a5f-af0a-4d4c-bd7b-62fcad0a3364', name='SiLA Service'),
'gen5_service': Gen5Service(client=<unitelabs.sdk.client.client.Client object at 0x7f5ed9101a50>, id='92e02f01-f877-44c0-bbcf-c225e3a9e56f', name='Gen5 Service'),
'door_controller': DoorController(client=<unitelabs.sdk.client.client.Client object at 0x7f5ed9101a50>, id='b536db1c-e343-4b64-a697-ea4d91ac6166', name='Door Controller'),
'simulation_controller': SimulationController(client=<unitelabs.sdk.client.client.Client object at 0x7f5ed9101a50>, id='ab8bb1b8-6a26-49f8-bace-5086b3d4a344', name='Simulation Controller'),
'temperature_controller': TemperatureController(client=<unitelabs.sdk.client.client.Client object at 0x7f5ed9101a50>, id='6c59aa90-740d-40ae-acae-ab8fa3e55aeb', name='Temperature Controller'),
'shake_controller': ShakeController(client=<unitelabs.sdk.client.client.Client object at 0x7f5ed9101a50>, id='416a0b31-fe5d-4fd8-9777-0c4ca569f483', name='Shake Controller'),
'protocol_controller': ProtocolController(client=<unitelabs.sdk.client.client.Client object at 0x7f5ed9101a50>, id='9db6a059-f693-4bc2-9ba0-0c9afe577ba0', name='Protocol Controller')}
Get Gen5 module actions
Get the actions (callable methods) associated with each module by calling gen5.<module>.actions.keys()
print(gen5.gen5_service.actions.keys())
dict_keys(['get_deactivate_plate', 'get_delete_active_plate', 'get_active_plate', 'get_available_protocols', 'get_column_count', 'get_data_set_names', 'get_device_connected', 'get_device_info', 'get_experiment_open', 'get_image_folder_path', 'get_modifiable_procedure', 'get_plate_calibration_type', 'get_plate_is_labware', 'get_plate_layout', 'get_plate_list', 'get_plate_types', 'get_procedure', 'get_protocol_path', 'get_read_in_progress', 'get_read_status', 'get_reader_status', 'get_row_count', 'get_sample_count', 'get_sample_description', 'get_version', 'get_vessel_count', 'abort_read', 'activate_plate', 'close_experiment', 'get_plate_data', 'new_experiment', 'resume_read', 'save_experiment', 'set_modifiable_procedure', 'set_plate_layout', 'set_procedure', 'set_protocol_path', 'start_read', 'validate_procedure'])
The Gen5 service module
The Gen5 service module inlcudes a lot of low level methods to get information, device status and data, start reads, get information about reads, the current protocol, and the plate.
Call basic reader info
# Get Gen5 version
await gen5.gen5_service.get_version()
'3.14.03'
# Check if the device is connected
await gen5.gen5_service.get_device_connected()
True
# Get the protocols that are available on the computer connected to the reader
await gen5.gen5_service.get_available_protocols()
[
'protocol.prt',
'protocol2.prt',
'Absorbance600nm.prt',
'ADH kinetic assay.prt',
]
# The path at which protocols are read from. This can be changed using gen5.gen5_service.set_protocol_path().
await gen5.gen5_service.get_protocol_path()
'C:\\Users\\Public\\Documents\\Protocols'
# Check if read in progress. Works always.
print(await gen5.gen5_service.get_read_in_progress())
# Check read status. Only works while the experiment is open and the plate is selected.
try:
print(await gen5.gen5_service.get_read_status())
except ExecutionError as error:
print(json.loads(error.message))
False
{'errorIdentifier': 'io.unitelabs/gen5/Gen5Service/v1/DefinedExecutionError/NoActivePlateError', 'message': 'None is not a valid active plate. Please make a new experiment and activate a plate before using this method.'}
Read a plate
To read a plate using a protocol created previously 3 steps are needed:
- Create a new experiment
- Select a plate
- Start the read
This section uses the following methods: close_experiment, new_experiment, activate_plate, get_current_active_plate, get_device_connected and start_read
gen5_service = gen5.gen5_service
# Close any open experiment. An open experiment will prevent a new one from being created.
# The close_experiment command does close the previous experiment without saving.
# For saving the experiment on the machine connected to the reader, the save_experiment() call can be used.
# It is advisable to utilize the get_plate_data() function to retrieve data and then save it manually or using Python
# calls, rather than using the save_experiment() method. Further details on how to implement this approach are
# provided later in this guide.
if (await gen5_service.get_experiment_open()):
await gen5_service.close_experiment()
print('Previous experiment closed without saving.')
# Create a new experiment that uses the "protocol" protocol. Absolut paths are possible, relative paths are relative to
# the protocol path (gen5_service.get_protocol_path())
# New protocols can be created in Gen5.
# Creating new protocols with UniteLabs SDK is partially possible (see "protocol controller")
await gen5_service.new_experiment(protocol='protocol.prt')
# Each experiment can hold multiple plates. The default is one plate, which means most of the time, activating the
# first plate in the experiment is sufficient.
await gen5_service.activate_plate(plate_name=(await gen5_service.get_plate_list())[0])
print('Active plate is: ', await gen5_service.get_active_plate()) # The standard name of the first plate is "Plate 1"
# If the device is connected, the read is started
if (await gen5_service.get_device_connected()):
await gen5_service.start_read()
Active plate is: Plate 1
# Get a list of all plates defined within the active experiment
await gen5.gen5_service.get_plate_list()
['Plate 1']
Read Status & Abort
If a read has been started, the status of said read can be checked by calling get_read_status().
This method returns a integer with the following meanings:
1: In progress
2: Aborted
3: Paused
4: Error
5: Completed
To abort a read, abort_read() is used. The connector will try to abort the read as soon as possible, but the reader sometimes blocks the abort for an undetermable amount of time.
await gen5_service.get_read_status()
1
await gen5_service.abort_read()
'Measurement aborted with final read status 2 (0: Not started, 1: In progress, 2: Aborted, 3: Paused, 4: Error, 5: Completed).'
Data sets
Depending on the used protocol, different data sets are available. To get the expected data set names for a plate, get_data_set_names() can be called (after a plate within an experiment was selected).
In this example the plate will be read using two wavelengths: $750,nm$ and $750,nm$, thus the two datasets to be expected are called 750 and 600. Dataset names starting with T° will not produce a data series on read and can be ignored for future implementations.
await gen5.gen5_service.get_data_set_names()
['750', '600', 'T° 750']
Getting and using the read data
A couple of example how to get and handle the data after starting a read.
For visual feedback, the following utility function plot_heatmap can be used.
# Plot data as heatmap (utility function):
def plot_heatmap(data, data_set_name):
try:
data = data[data_set_name]
except KeyError:
print(f'No data found for {data_set_name}')
return
max_row = max(max(row) for row in data["row"]) + 1
max_col = max(max(col) for col in data["column"]) + 1
grid = np.zeros((max_row, max_col))
for row, col, val in zip(data["row"], data["column"], data["value"]):
for r, c, v in zip(row, col, val):
grid[r, c] = v
plt.figure(figsize=(6, 5))
plt.imshow(grid, cmap='hot', interpolation='nearest')
plt.title(f"Active Read ({data_set_name})")
plt.colorbar()
plt.grid(which='both', color='white', linestyle='-', linewidth=0.5)
plt.xticks(np.arange(max_col))
plt.yticks(np.arange(max_row))
plt.show()
The setup to start a new read with the protocol.prt as explained before:
protocol = 'protocol.prt'
gen5_service = gen5.gen5_service
if (await gen5_service.get_experiment_open()):
await gen5_service.close_experiment()
print('Previous experiment closed without saving.')
await gen5_service.new_experiment(protocol=protocol)
await gen5_service.activate_plate(plate_name=(await gen5_service.get_plate_list())[0])
if (await gen5_service.get_device_connected()):
await gen5_service.start_read()
Previous experiment closed without saving.
Read the data (1)
The data is buffered within the connector until a new read is started. This means however the data is accessed, it is temporarily available with the get_plate_data() call until a new read is started (can / needs to be called multiple times).
The get_plate_data() updates the buffered data one line at a time. This means it needs to be called multiple times until no more updates are happening. Within the data there is a current_data_status integer (data["current_data_status"]), which can be used to check if more data is available:
0: No raw data is currently available.
1: Raw data was found. More data is available.
2: Raw data was found. No additional data is currently available.
There are a couple of ways to read the produced data.
- Wait until the read is concluded
a. Print the data to the command line
b. Write the data to a json file
c. Save the different wavelengths reads to multiple csv files
d. Display and save as a heatmap
# 1. To wait until the read is concluded use:
while (await gen5_service.get_read_status()) != 5:
clear_output()
print('Waiting')
await asyncio.sleep(2)
print('Read concluded')
# Now call get_plate_data until no more data is found:
# This is necessary as get_plate_data reads one line at a time.
data = json.loads(await gen5_service.get_plate_data())
while (data["current_data_status"] == 1): # 0: No raw data is currently available, 1: Raw data was found. More data is available., 2: Raw data was found. No additional data is currently available.
data = json.loads(await gen5_service.get_plate_data())
# a. print the data
print(data)
# b. save the whole data as .json file:
with open('output_raw.json', 'w') as file:
json.dump(data, file)
# c. save only values as CSVs:
for dataset, content in data.items(): # loop through all the wavelength measured
if dataset == "current_data_status":
continue
max_row = max(max(row) for row in content['row'])
max_col = max(max(col) for col in content['column'])
csv_matrix = [['' for _ in range(max_col + 1)] for _ in range(max_row + 1)]
for row, col, val in zip(content['row'], content['column'], content['value']):
for r, c, v in zip(row, col, val):
csv_matrix[r][c] = v
with open(f'output_{dataset}.csv', 'w', newline='') as file:
writer = csv.writer(file)
writer.writerows(csv_matrix)
# d. display plate as heatmap:
for dataset in data.keys():
if dataset == "current_data_status":
continue
plot_heatmap(data, dataset)
Waiting
Read concluded
{'750': {'status': [], 'row': [[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1], [2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2], [3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3], [4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4], [5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5], [6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6], [7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7]], 'column': [[0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11], [11, 10, 9, 8, 7, 6, 5, 4, 3, 2, 1, 0], [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11], [11, 10, 9, 8, 7, 6, 5, 4, 3, 2, 1, 0], [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11], [11, 10, 9, 8, 7, 6, 5, 4, 3, 2, 1, 0], [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11], [11, 10, 9, 8, 7, 6, 5, 4, 3, 2, 1, 0]], 'kineticIndex': [[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]], 'wavelengthIndex': [[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]], 'horizontalIndex': [[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]], 'verticalIndex': [[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]], 'value': [[-0.0, -0.0, 0.0, -0.0, -0.0, 0.0, 0.0, 0.0, -0.0, -0.0, -0.0, 0.0], [-0.0, 0.0, -0.0, -0.0, -0.0, 0.0, -0.0, -0.0, -0.0, -0.0, -0.0, -0.0], [0.0, 0.0, -0.0, -0.0, -0.0, -0.0, -0.0, -0.0, -0.0, 0.0, -0.0, 0.0], [-0.0, 0.0001, -0.0, -0.0, 0.0, -0.0, 0.0, -0.0, -0.0, -0.0, -0.0, -0.0], [-0.0, -0.0, -0.0, 0.0, -0.0, -0.0, -0.0, 0.0, -0.0, -0.0, -0.0, 0.0], [-0.0, 0.0, -0.0, -0.0, -0.0, -0.0, 0.0, 0.0, -0.0, -0.0, -0.0, -0.0], [-0.0, -0.0, -0.0, -0.0, -0.0, 0.0, -0.0, -0.0, -0.0, -0.0, 0.0, -0.0], [-0.0, -0.0, -0.0, -0.0, 0.0, -0.0, 0.0, -0.0, -0.0, -0.0, 0.0, 0.0]], 'primaryStatus': [[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]], 'secondaryStatus': [[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]]}, 'current_data_status': 2, '600': {'status': [], 'row': [[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1], [2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2], [3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3], [4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4, 4], [5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5], [6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6, 6], [7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7]], 'column': [[0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11], [11, 10, 9, 8, 7, 6, 5, 4, 3, 2, 1, 0], [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11], [11, 10, 9, 8, 7, 6, 5, 4, 3, 2, 1, 0], [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11], [11, 10, 9, 8, 7, 6, 5, 4, 3, 2, 1, 0], [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11], [11, 10, 9, 8, 7, 6, 5, 4, 3, 2, 1, 0]], 'kineticIndex': [[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]], 'wavelengthIndex': [[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]], 'horizontalIndex': [[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]], 'verticalIndex': [[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]], 'value': [[0.0001, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0001, -0.0, 0.0001, -0.0, 0.0, -0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, -0.0, 0.0, 0.0, 0.0], [0.0, -0.0, 0.0001, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, -0.0], [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, -0.0, -0.0, 0.0, -0.0], [0.0, 0.0, 0.0001, 0.0, -0.0, 0.0, -0.0, 0.0, 0.0, 0.0, 0.0001, 0.0], [0.0, 0.0, -0.0, 0.0, 0.0, 0.0001, 0.0, 0.0001, 0.0, 0.0, 0.0, 0.0001], [-0.0, 0.0001, 0.0, -0.0, 0.0, 0.0, 0.0, 0.0, 0.0001, 0.0001, 0.0, 0.0], [-0.0, 0.0, 0.0, 0.0, 0.0, 0.0, -0.0, 0.0, 0.0001, 0.0, 0.0, 0.0001]], 'primaryStatus': [[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]], 'secondaryStatus': [[0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0]]}}
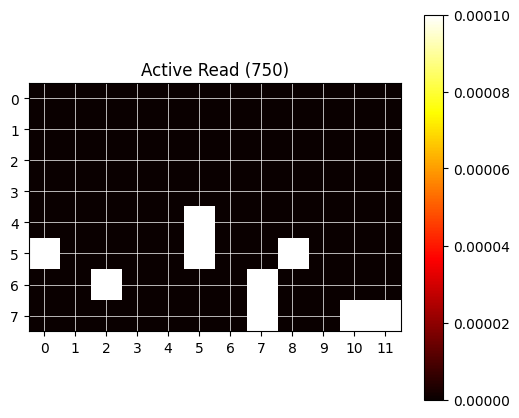
Read the data (2)
- Read the data during the read (live)
The heatmap is updated each time new data is available
# As long as the read is running, we gather the result data.
data = {}
while (await gen5_service.get_read_status()) == 1:
try:
new_data = json.loads(await gen5_service.get_plate_data())
if new_data["current_data_status"] != 0 and data != new_data:
data = new_data
# perform one of the options shown above. For example plot the heatmap of the first dataset:
data_set_names = await gen5_service.get_data_set_names()
clear_output()
for data_set_name in data_set_names:
plot_heatmap(data, data_set_name)
await asyncio.sleep(1)
except Exception as e:
print(e)
print(data)
# when the read is concluded, there might still be leftover data saved on the plate.
# The same lines as shown in the previous section can be used to collect all of this data:
data = json.loads(await gen5_service.get_plate_data())
data_set_names = await gen5_service.get_data_set_names()
while (data["current_data_status"] == 1):
data = json.loads(await gen5_service.get_plate_data())
clear_output()
for data_set_name in data_set_names:
plot_heatmap(data, data_set_name)
No data found for T° 750
Door controller (= carrier controller)
The door_controller module contains properties to monitor and commands to controll the carrier of the reader:
- subscribe_door_open()
- get_door_open_once()
- close_door()
- open_door()
Subscribe to the door state
A door state of True indicates an open door (carrier outside) while False indicates a closed door (carrier inside)
from contextlib import suppress
subscription = await gen5.door_controller.subscribe_door_open()
try:
with suppress(asyncio.CancelledError):
async with subscription:
async for item in subscription:
print(item)
except KeyboardInterrupt as e:
print("Stopped by user")
False
True
Toggle the door
Here a simple door toggle mechanism is shown.
# Example: a carrier toggle:
if (await gen5.door_controller.get_door_open_once()):
await gen5.door_controller.close_door()
else:
await gen5.door_controller.open_door()
print(await gen5.door_controller.get_door_open_once())
False
Temperature Controller
The temperature_controller module contains properties to monitor and commands to controll the temperature set point:
- subscribe_current_temperature()
- get_target_temperature()
- set_target_temperature(temperature in °C)
- get_incubation_on()
- turn_incubation_on()
- turn_incubation_off()
from contextlib import suppress
subscription = await gen5.temperature_controller.subscribe_current_temperature()
try:
with suppress(asyncio.CancelledError):
async with subscription:
async for current_temperature in subscription:
clear_output()
print(f"Current temperature: {round(current_temperature, 2)}°C")
except KeyboardInterrupt as e:
print("Stopped by user")
Current temperature: 24.8°C
Example implementation for demonstration
In this implementation: a. set a target temperature b. turn incubation on c. monitor the temperature change with a subscription d. stop the monitoring as soon as target temperature is reached e. turn incubation off
target_temperature = 25
# a
await gen5.temperature_controller.set_target_temperature(target_temperature=target_temperature)
# b
await gen5.temperature_controller.turn_incubation_on()
# c
subscription = await gen5.temperature_controller.subscribe_current_temperature()
try:
with suppress(asyncio.CancelledError):
async with subscription:
async for current_temperature in subscription:
clear_output()
print(f"Current temperature: {round(current_temperature, 2)}°C")
# d
if current_temperature >= target_temperature:
print(f"Target temperature of {round(target_temperature, 2)}°C reached.")
break
except KeyboardInterrupt:
print("Stopped by user")
# e
await gen5.temperature_controller.turn_incubation_off()
Current temperature: 25.0°C
Target temperature of 25°C reached.
Shaker controller
The shaker controller has three calls to perform a simple shaker step, monitor it's state and abort it:
- start_shake_step(mode,duration_sec,displacement_mm,orbital_speed)
- abort_shake
- get_shake_in_progress
The abort_shake and get_shake_in_progress are the same implementation as the abort_read and get_read_in_progress, as the internal mechanism does not record the difference between these steps. This means that a abort_shake call will also abort a read if one is in progress.
if (await gen5_service.get_experiment_open()):
await gen5_service.close_experiment()
print('Previous experiment closed without saving.')
await gen5.shake_controller.start_shake_step(mode=0,duration_sec=60.0,displacement_mm=2,orbital_speed=0)
Previous experiment closed without saving.
await gen5.shake_controller.abort_shake()
'Measurement aborted with final read status 2 (0: Not started, 1: In progress, 2: Aborted, 3: Paused, 4: Error, 5: Completed).'
Protocol controller
The protocol controller offers the automated creation of a protocol with only absorption steps and shake steps. This controller also allows to change the plate type to a predifined plate type within Gen5.
All the actions that do not change the protocol itself (plate editing, protocol information getters) also work for protocols previously designed in Gen5.
gen5_pc = gen5.protocol_controller
gen5_service = gen5.gen5_service
Initialize an empty protocol
The first step to create a protocol is to call initialize_empty_protocol() (if there is already a protocol open, it needs to be closed first)
if (await gen5_service.get_experiment_open()):
await gen5_service.close_experiment()
print('Previous experiment closed without saving.')
await gen5_pc.initialize_empty_protocol()
Previous experiment closed without saving.
Get and set steps
The get_step_list() call of the protocol controller can be used to inspect all the steps currently planned in the protocol. This also works for previously defined protocols.
The empty protocol which was just initialized returns an empty list as no steps have been defined.
await gen5_pc.get_step_list()
[]
Create an absorbance step
The add_absorbance_step method requires the following settings:
- index: integer; the index at which the step should be placed
- wavelength: string; of comma separated integers of the wavelengths to be read
- read_speed: integer; the speed of the read step (0: normal, 1: sweep)
- wavelength_switching: boolean; whether to switch wavelengths per well. Only if multiple wavelengths are provided.
await gen5_pc.add_absorbance_step(index=0, wavelengths="450,700", read_speed=1, wavelength_switching=True)
Now get_step_list() returns a list with a single ReadStep.
The details of a step can be read by using get_step(index).
print(await gen5_pc.get_step_list())
print('--------')
print(await gen5_pc.get_step(index=0))
['ReadStep']
--------
<ReadStep>
<Detection>Absorbance</Detection>
<ReadType>EndPoint</ReadType>
<Wells>Full Plate</Wells>
<Measurements>
<Measurement Index="1">
<Wavelength>450</Wavelength>
</Measurement>
<Measurement Index="2">
<Wavelength>700</Wavelength>
</Measurement>
</Measurements>
<ReadSpeed>Sweep</ReadSpeed>
<DelayAfterPlateMovementMSec>100</DelayAfterPlateMovementMSec>
<MeasurementsPerDataPoint>8</MeasurementsPerDataPoint>
</ReadStep>
Create an shake step
The add_shake_step method requires the following settings:
- index: integer; the index of the shake step.
- mode: integer; the mode of the shake step (0: linear, 1: orbital or 2: double orbital).
- duraction_sec: float; the duration of the shake step in seconds.
- displacement_mm: integer; the displacement of the shake step in millimetres.
- orbital_speed: integer; the orbital speed of the shake step (0: slow, 1: fast). Only for orbital and double orbital mode.
await gen5_pc.add_shake_step(index=0, mode=0, duration_sec=60.0, displacement_mm=2, orbital_speed=1)
Now get_step_list() returns a list with a two steps.
Note that because the shake step was also added at index 0, the read step was pushed back.
To delete a step delete_step(index) is available.
await gen5_pc.get_step_list()
['ShakeStep', 'ReadStep']
await gen5_pc.delete_step(index=0)
await gen5_pc.get_step_list()
['ReadStep']
Change plate type
The plate to be read can be changed using the get_plate_type() call.
The default plate of an "empty_protocol" is a 96 WELL PLATE.
The plate type can only be changed to plate types defined within Gen5.
To check the available types get_supported_plate_types() can be used (note: in the example below [:10] is used to only print the first 10).
This also works for protocols previously designed in Gen5.
await gen5_pc.get_plate_type()
'96 WELL PLATE, Lid: true'
await gen5_pc.set_plate_type(plate_type='24 WELL PLATE', use_lid=False)
print(await gen5_pc.get_plate_type())
print('\n\n Defined plates (10 examples):')
(await gen5_pc.get_supported_plate_types())[:10]
24 WELL PLATE, Lid: false
Defined plates (10 examples):
['24 WELL PLATE',
'48 WELL PLATE',
'96 WELL PLATE',
'384 WELL PLATE',
'72 WELL PLATE TERASAKI',
'60 WELL PLATE TERASAKI',
'96 WELL PLATE TERASAKI',
'96 WELL PLATE HELMA',
'6 WELL PLATE',
'12 WELL PLATE']
Run the newly created protocol
The newly created protocol (here with one read step using two wavelength and a 24 well plate) can now be run and observed using the previosly written code (see The Gen5 service module>Read the data (2)).
await gen5_service.activate_plate(plate_name=(await gen5_service.get_plate_list())[0])
if (await gen5_service.get_device_connected()):
await gen5_service.start_read()
# As long as the read is running, we gather the result data.
data = {}
while (await gen5_service.get_read_status()) == 1:
try:
new_data = json.loads(await gen5_service.get_plate_data())
if new_data["current_data_status"] != 0 and data != new_data:
data = new_data
# perform one of the options shown above. For example plot the heatmap of the first dataset:
data_set_names = await gen5_service.get_data_set_names()
clear_output()
for data_set_name in data_set_names:
plot_heatmap(data, data_set_name)
await asyncio.sleep(1)
except Exception as e:
print(e)
print(data)
# when the read is concluded, there might still be leftover data saved on the plate.
# The same lines as shown in the previous section can be used to collect all of this data:
data = json.loads(await gen5_service.get_plate_data())
data_set_names = await gen5_service.get_data_set_names()
while (data["current_data_status"] == 1):
data = json.loads(await gen5_service.get_plate_data())
clear_output()
for data_set_name in data_set_names:
plot_heatmap(data, data_set_name)
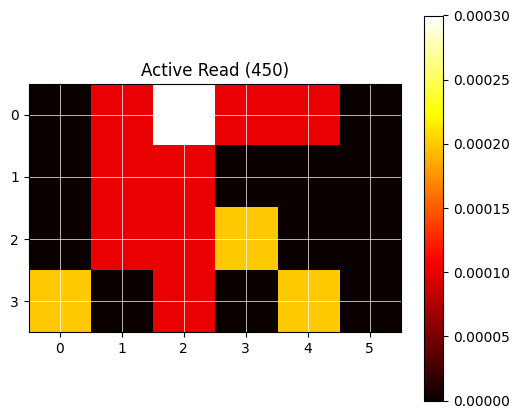
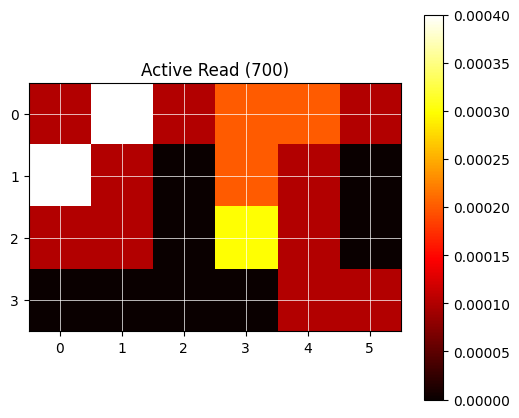
No data found for T° 450
Command set
Refere to this if you want to use an action that was not (sufficiently) explained in the examples above.
Other helpful calls to explor actions:
- Type of action (Property, Sensor or Controll):
print(await gen5.<module>.<action>.type) - Parameters:
print(await gen5.<module>.<action>.parameters) - Returns:
print(await gen5.<module>.<action>.responses)
Note:
- A PROPERTY returns a response and doesn't require any parameters.
- A SENSOR returns a subscription of responses and doesn't require parameters.
- A CONTROL requires parameters and can return a single response or a subscription of responses.
gen5_service
gen5.gen5_service.get_version()
Get the version of the installed Gen5 software.
gen5.gen5_service.get_device_connected()
Test the connection to the connected reader device. Returns false if in simulation mode.
gen5.gen5_service.get_plate_types()
Return a list of all available plate names.
gen5.gen5_service.get_device_info()
Return a JSON string with information about the connected device:
{ "eAbsorbanceReadSupported": bool, "eAbsorbanceWavelengthMin": int, "eAbsorbanceWavelengthMax": int, "eAbsorbanceReadModeCount": int, "eAbsorbanceReadModeName": dict, "eTemperatureControlOption": [str], "eTemperatureMin": int, "eTemperatureMax": int, "eTemperatureGradientMax": int, "eShakeSupported": bool, "eSerialNumber": str, "eInstrumentName": str, "eFilterFluorescenceSupported": bool, "eMonoFluorescenceSupported": bool, "eReaderArchitectureLevel": int }
gen5.gen5_service.get_reader_status()
Return the status of the reader.
gen5.gen5_service.get_available_protocols()
Return a list of available protocols.
Potential errors:
- PathNotFoundError
- DBError
gen5.gen5_service.new_experiment(protocol: str)
Create a new experiment.
Parameters:
protocol: The name of the protocol (must be available in the list of available protocols).Potential errors:
- ExperimentAlreadyOpenError
gen5.gen5_service.get_experiment_open()
Return True if an experiment is currently open.
gen5.gen5_service.save_experiment(path: str, name: str)
Save the current experiment to the given path.
Parameters:
path: The path to save the experiment to.name: The file name of the experiment.Returns:
- A confirmation message.
Potential errors:
- NoExperimentError
gen5.gen5_service.close_experiment()
Close the current experiment without saving.
Potential errors:
- NoExperimentError
gen5.gen5_service.get_plate_layout()
Returns the layout of the plate in a BTIPlateLayout XML format.
Potential errors:
- NoActivePlateError
- NoExperimentError
gen5.gen5_service.set_plate_layout(layout: str)
Set the layout of the plate.
Parameters:
layout: The layout of the plate in a BTIPlateLayout XML format.Potential errors:
- NoActivePlateError
- NoExperimentError
gen5.gen5_service.get_protocol_path()
Return the path of the protocol folder.
gen5.gen5_service.set_protocol_path(path: str)
Sets the path to the protocol folder. Doesn't remember if connector is restarted.
Parameters:
path: The absolute path where the .prt files are located.Potential errors:
- PathNotFoundError
gen5.gen5_service.get_plate_list()
A list of names of available plates.
Potential errors:
- NoExperimentError
gen5.gen5_service.activate_plate(plate_name: str)
Activate a plate for the current experiment.
Parameters:
plate_name: The name of the plate to activate (must be in the list of available plates).Potential errors:
- NoExperimentError
- PlateNotFoundError
gen5.gen5_service.deactivate_plate()
Deactivate the currently active plate.
Potential errors:
- NoActivePlateError
gen5.gen5_service.get_active_plate()
Return the name of the currently active plate.
Potential errors:
- NoActivePlateError
gen5.gen5_service.delete_active_plate()
Delete the currently active plate. (Not deactivating, deletes plate and all data!)
Potential errors:
- NoActivePlateError
gen5.gen5_service.get_procedure()
Return the current procedure.
Potential errors:
- NoActivePlateError
gen5.gen5_service.get_modifiable_procedure()
Return the part of the current procedure, which can be modified.
Potential errors:
- NoActivePlateError
gen5.gen5_service.set_procedure(procedure: str)
Set the procedure for the current experiment.
Parameters:
procedure: The procedure to set.Potential errors:
- NoActivePlateError
- ReadInProgressError
gen5.gen5_service.set_modifiable_procedure(procedure: str)
Set the modifiable part of the procedure.
Parameters:
procedure: The modifiable part of the procedure to set.Potential errors:
- NoActivePlateError
- ReadInProgressError
gen5.gen5_service.validate_procedure(flip: bool)
Validate the current procedure.
Parameters:
flip: Whether to flip the plate for validation.Returns:
- A validation status (1: OK, any other number: Error Code).
- A validation message.
Potential errors:
- NoActivePlateError
- ReadInProgressError
- MethodNotSupportedError
gen5.gen5_service.start_read()
Start reading the active plate.
Potential errors:
- NoActivePlateError
- ReadInProgressError
- ReadStartError
gen5.gen5_service.resume_read()
Resume the current read.
Potential errors:
- NoActivePlateError
- NoActiveReadError
gen5.gen5_service.abort_read()
Abort the current read.
Returns:
- A confirmation message that the read was aborted.
Potential errors:
- NoActivePlateError
- NoActiveReadError
gen5.gen5_service.get_read_status()
Return the status of the current read.
Status codes:
- 0: Not started
- 1: In progress
- 2: Aborted
- 3: Paused
- 4: Error
- 5: Completed
Potential errors:
- NoActivePlateError
gen5.gen5_service.get_plate_calibration_type()
Return the calibration type of the active plate. Can be either Standard Plate (0) or Calibration Plate (1).
Potential errors:
- NoActivePlateError
gen5.gen5_service.get_sample_count()
Return the number of samples assigned to the current plate. This property only applies to experiments based on Custom or Open Protocols.
Note: This property is not applicable to panels that have unique samples for each protocol.
Potential errors:
- NoActivePlateError
gen5.gen5_service.get_row_count()
Return the number of rows in the active plate.
Potential errors:
- NoActivePlateError
gen5.gen5_service.get_column_count()
Return the number of columns in the active plate.
Potential errors:
- NoActivePlateError
gen5.gen5_service.get_plate_is_labware()
Return True if the active plate is labware.
Potential errors:
- NoActivePlateError
gen5.gen5_service.get_vessel_count()
Return the number of vessels for the current plate type. If the plate is not labware the vessel count = 1.
Potential errors:
- NoActivePlateError
gen5.gen5_service.get_data_set_names()
Return a list of available data set names.
Potential errors:
- NoActivePlateError
gen5.gen5_service.get_plate_data()
Return the topmost entry in the data stack. Must be called more than once to retrieve all data.
Returns:
- A JSON (string) of the data:
{ "str dataset_name (f.ex. wavelength)": { "row": [[int]]; An array of longs (int) containing the row (0-based) of the current read, "column": [[int]]; An array of longs (int) containing the column (0-based) of the current read, "value": [[float]]; An array of doubles (float) containing the value of the current read, "kineticIndex": [[int]]; An array of longs (int) containing the kinetic index (0-based) of the current read, "wavelengthIndex": [[int]]; An array of longs (int) containing the spectrum wavelength index (0-based) of the current read, "horizontalIndex": [[int]]; An array of longs (int) containing the horizontal index (0-based) of the current read, "verticalIndex": [[int]]; An array of longs (int) containing the vertical index (0-based) of the current read, "primaryStatus": [[int]]; An array of longs (int) containing the primary status of the current read, ( 0: The well was successfully read, 1: The well data was outside the reader's range, 2: The reader was not able to capture the well data, 3: The well was masked (read from file only), 4: An error was encountered reading the well, 5: The well was not read ) "secondaryStatus": [[int]]; An array of longs (int) containing the secondary status of the current read, ( 0: There are no secondary status conditions, 1: The well was being dispensed with injector 1 during the read, 2: The well was being dispensed with injector 2 during the read, 3: The well was being dispensed with injector 3 during the read, 4: The well was being dispensed with injector 4 during the read ) }, "600": { "row": [[0, 0, 0], [1, 1, 1], [2, 2, 2]], "column": [[0, 1, 2], [0, 1, 2], [0, 1, 2]], "value": [[0.1, 0.2, 0.3], [0.4, 0.5, 0.6], [0.7, 0.8, 0.9]], ... } }Potential errors:
- NoActivePlateError
gen5.gen5_service.get_sample_description()
Return a dictionary with the sample description.
Potential errors:
- NoActivePlateError
gen5.gen5_service.get_image_folder_path()
Return the count and the list of image folders that apply to the current plate.
Potential errors:
- NoActivePlateError
gen5.gen5_service.get_read_in_progress()
Return True if a read is currently in progress.
door_controller
gen5.door_controller.subscribe_door_open()
Subscribe to the state of the door.
gen5.door_controller.get_door_open_once()
Retrieve the door state once.
gen5.door_controller.open_door(status: sila.Status, intermediate: sila.Intermediatestr)
The abstract method to open the door.
Yields:
- The current status of the door opening process as a string.
Returns:
- The state of the door where true means open.
- The end status of the door as a string.
Potential errors:
- ReadInProgressError
gen5.door_controller.close_door(status: sila.Status, intermediate: sila.Intermediatestr)
The abstract method to close the door.
Yields:
- The current status of the door opening process as a string.
Returns:
- The state of the door where true means open.
- The end status of the door as a string.
Potential errors:
- ReadInProgressError
temperature_controller
gen5.temperature_controller.subscribe_current_temperature()
Returns the currently measured temperature in °C.
gen5.temperature_controller.get_target_temperature()
Returns the currently set target temperature in °C.
gen5.temperature_controller.set_target_temperature(target_temperature: int)
Set the target temperature in °C.
Parameters:
target_temperature: The target temperature to set in °C.
shake_controller
gen5.shake_controller.start_shake_step(mode: int, duration_sec: float, displacement_mm: int, orbital_speed: int)
Start a shake step with the given parameters. This creates a new experiment with only the shake step. Any previous experiment has to be closed before calling this method. Closing an experiment will discard all unsaved data of the experiment.
Parameters:
mode: The mode of the shake step (0: linear, 1: orbital, 2: double orbital).duration_sec: The duration of the shake step in seconds.displacement_mm: The displacement of the shake step in millimeters.orbital_speed: The orbital speed of the shake step (0: slow, 1: fast). Only for orbital and double orbital mode.Potential errors:
- ReadInProgressError
- ExperimentAlreadyOpenError
- InvalidParameterError
- ShakeStartError
gen5.shake_controller.abort_shake()
Abort the current read step. Shaking is considered a read step. Is the same as gen5_service.abort_read().
Returns:
- A confirmation message that the read was aborted.
Potential errors:
- NoActivePlateError
- NoActiveShakeError
gen5.shake_controller.get_shake_in_progress()
Return True if a read is currently in progress. Shaking is considered a read in progress. Does the same as gen5_service.get_read_in_progress().
protocol_controller
gen5.protocol_controller.initialize_empty_protocol()
Initializes an empty protocol. Potential errors: ReadInProgressError, ExperimentAlreadyOpenError
gen5.protocol_controller.get_step_list()
Returns the list of steps in the current protocol.
gen5.protocol_controller.get_step(index: int)
Returns the step at the given index.
Parameters:
index: The index of the step to return.Returns:
- The step at the given index.
gen5.protocol_controller.delete_step(index: int)
Deletes the step at the given index.
Parameters:
index: The index of the step to delete.
gen5.protocol_controller.add_absorbance_step(index: int, wavelengths: str, read_speed: int, wavelength_switching: bool)
Start an absorbance step with the given parameters. This creates a new experiment with only the absorbance step. Any previous experiment has to be closed before calling this method. Closing an experiment will discard all unsaved data of the experiment.
Parameters:
index: The index of the absorbance step.wavelengths: Wavelength(s) to measure. For multiple wavelengths, use comma-separated values.read_speed: The speed of the read step (0: normal, 1: sweep).wavelength_switching: Whether to switch wavelengths per well. Only if multiple wavelengths are provided.
gen5.protocol_controller.add_shake_step(index: int, mode: int, duration_sec: float, displacement_mm: int, orbital_speed: int)
Start a shake step with the given parameters. This creates a new experiment with only the shake step. Any previous experiment has to be closed before calling this method. Closing an experiment will discard all unsaved data of the experiment.
Parameters:
index: The index of the shake step.mode: The mode of the shake step (0: linear, 1: orbital, 2: double orbital).duration_sec: The duration of the shake step in seconds.displacement_mm: The displacement of the shake step in millimeters.orbital_speed: The orbital speed of the shake step (0: slow, 1: fast). Only for orbital and double orbital mode.
gen5.protocol_controller.get_plate_type()
Returns the type of the plate.
gen5.protocol_controller.get_supported_plate_types()
Returns the supported plate types.
gen5.protocol_controller.set_plate_type(plate_type: str, use_lid: bool)
Sets the type of the plate.
Parameters:
plate_type: The type of the plate. This has to be one of the supported plate types.use_lid: Whether to use a lid.